Parsing Common Crawl in a day for $60
# December 14, 2023
Common Crawl is the largest permissively licensed archive of the Internet. They conduct their crawls about 5 times a year and index about 3.5 billion pages every crawl.
In addition to forming a bulk of the foundation of modern language models, there's a ton of other data buried within Common Crawl. Incoming and external links to websites, referral codes, leaked data. If it's public on the Internet, there's a good chance CC has it somewhere within its index. So much critical data is just sitting there, buried somewhere in the html markup. 1
It's a dataset that I keep coming back to. And I've never been particularly happy with my method of querying it. Back when my previous jobs had a big Spark cluster, I'd typically use that - or sometimes Hadoop or sometimes even Athena. But inevitably managing these systems becomes a headache, prone to errors or crashes halfway through. Not to mention it's expensive and you still have to let the job queue run overnight.
I wanted to process one full revision of common crawl in a day, from scratch, and make it work robustly. That meant one machine and no distributed computation packages. Most libraries with this goal focus on how to process the index files. Index files are much distilled records that just have basic attributes about each page - the URL, language, page headers, etc. Since these files are indexed as parquet files they can be queried much more performantly via OLAP databases like duckdb. Here, on the other hand, I needed to do the whole HTML enchilada.
WAT is WARC? And for that matter, WET?
There are three file formats in each Common Crawl.
- WAT: Metadata files. Contains the success codes, language of the page, etc.
- WET: Plaintext extractions from the html. The equivalent of doing a
BeautifulSoup(html).get_text() - WARC: Raw crawl content. Contains the html files that were seen at crawl time and a few of the headers that are also in the WAT metadata files.
These data sizes grow as we get closer to the raw source. They all have 80,000 files but WARC weighs in at 86.77TB compressed whereas WET files are only 8.69TB.
Language of choice
Python has some well-supported parsers for warc files. I've used these before in Spark and they work pretty well but always did feel a bit sluggish. Throughput here is of the upmost importance if we're limited to some non-infinite number of CPU cores. Furthermore, Python's multiprocessing package works well for shorter running jobs, but I've always encountered strange crashes or inter-process communication errors when running these longer jobs.
I wanted to go for something a bit more battle tested at scale.
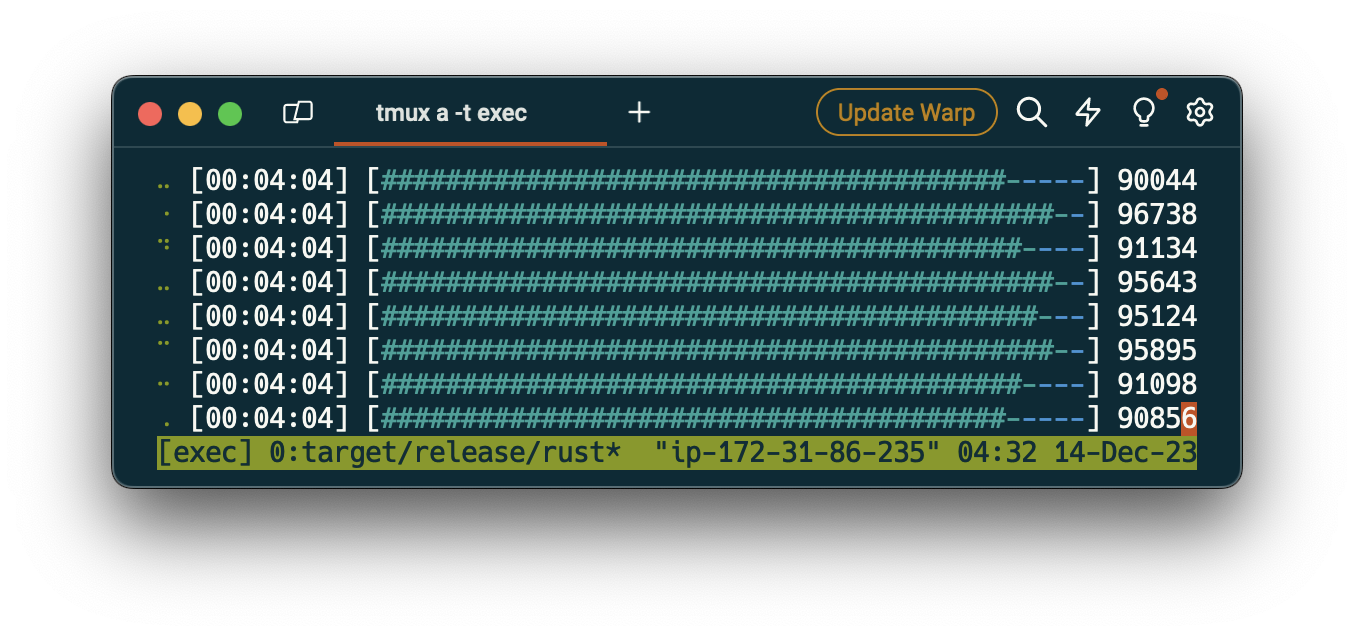
I wrote two POC parsers to see if there's one language that's a standout here for parsing the warc files from disk to memory. For Go I used a slyrz/warc and in Rust jedireza/warc. These are both parsing the same gzipped file that's already been downloaded on disk.
Go
First file URL: crawl-data/CC-MAIN-2023-40/segments/1695233505362.29/warc/
CC-MAIN-20230921073711-20230921103711-00000.warc.gz
114964 [________________________________________________________] ?% 4117 p/s
Processing completed in 28.128146833s. Found 22365 forms.
Rust
Finished release [optimized] target(s) in 1.60s
Running `target/release/rust`
First file URL: crawl-data/CC-MAIN-2023-40/segments/1695233505362.29/warc/
CC-MAIN-20230921073711-20230921103711-00000.warc.gz
[00:00:25] [████████████████████████████████████████] 0/0 Processing complete
Processing completed in 25.389549583s. Found 27983 forms.
These results were relatively stable over multiple executions. Rust barely edges out over Golang but not to a particularly meaningful level. I suspect this is near the theoretical maximum throughput on my Macbook Pro.
Once the records are in memory, they need to be processed. Which library has faster primitives to use? Which has better library support? That comes down heavily to what you're trying to do. For simple regex searches, both languages are again on parity. But in my case I also needed to implement a filter based on the page language to isolate English-only pages2. In an informal local benchmark, whatlang-rs beat out the other options available in both Rust and Go by a pretty wide margin while still keeping result accuracy pretty high. I picked Rust.
Filtering per datapoint
There are 86.77TB worth of compressed html files within the latest crawl. In an O(N) setting where the N is this big, you need to reduce your processing per datapoint. Every millisecond really does count. Ideally you can also get your average case to as close to a jump as possible.
I'll bold this for emphasis: Your filtering logic should be as dumb as possible.
It's hard to boil the ocean with a ton of logic and overhead processing for each page. Instead, consider the minimum metadata that you'll need to analyze before deciding if the page is relevant to your task:
- Presence or absence of some text
- Outgoing links
- Page language
- Current URL
This will become your filter logic that quickly validates or invalidates a page, queuing it for additional heavier work if needed. You should generally structure this filter as a text matching task. Conducting full HTML serialization, no matter how fast your underlying library, is going to prove a big bottleneck over those billions of iterations.
Regex is a good default for these pipelines. Compiled regexes are usually pretty fast and you have full expressiveness of a regular language. But sometimes implementing a tokenizer is going to be more efficient. Let's consider stripping html into raw text.
pub fn strip_nontext_regex(content: &str) -> String {
lazy_static! {
static ref CLEAN_HTML_REGEX: Regex = Regex::new(r#"<[^>]*>"#).unwrap();
static ref CLEAN_SCRIPTS_REGEX: Regex = Regex::new(r#"<script[^>]*>.*?</script>|<style[^>]*>.*?</style>"#).unwrap();
}
// Most specific
let raw_text = CLEAN_SCRIPTS_REGEX
.replace_all(&content, "")
.into_owned();
// Least specific
let raw_text = CLEAN_HTML_REGEX
.replace_all(&raw_text, "")
.into_owned();
raw_text
}
This was my original code to strip tags from the body to do some additional language detection. Two pretty straightforward regexes to clear out <tag> wrappers and everything inside of <script> and <style> tags.
However, this benchmarks to nearly 1000µs (0.001s). Across 3 billion pages that's 3 million seconds (833 hours) just for tag stripping. That's not to mention the language detection or data extraction.
Running benches/parse_benchmark.rs (target/release/deps/parse_benchmark-e83e2b63285db56d)
Gnuplot not found, using plotters backend
Benchmarking My Group/strip-text: stack: Warming up for 3.00Benchmarking
My Group/strip-text: regex
time: [936.51 µs 938.93 µs 941.85 µs]
Found 5 outliers among 100 measurements (5.00%)
3 (3.00%) high mild
2 (2.00%) high severe
If you refactor into a lexer, you'll see some pretty meaningful performance gains:
pub fn strip_nontext(content: &str) -> String {
let mut result = String::new();
let mut in_tag = false;
let mut script_or_style_block = false;
let mut index = 0;
let mut chars = content.chars().peekable();
while let Some(c) = chars.next() {
match c {
'<' => {
in_tag = true;
if let Some(next_char) = chars.peek() {
if *next_char == 's' || *next_char == 'S' {
// Look ahead to check for script or style
let lookahead = &content[index..];
if starts_with_lower(lookahead, "<script") || starts_with_lower(lookahead, "<style") {
script_or_style_block = true;
}
}
}
},
'>' if in_tag => {
in_tag = false;
let lookbehind = &content[..index];
if script_or_style_block && (ends_with_lower(lookbehind, "</script") || ends_with_lower(lookbehind, "</style")) {
script_or_style_block = false;
}
},
_ if !in_tag && !script_or_style_block => {
result.push(c);
},
_ => {}
}
index += c.len_utf8();
}
result
}
My Group/strip-text: stack: Collecting 100 sampMy Group/strip-text: stack
time: [478.61 µs 479.92 µs 481.27 µs]
Found 5 outliers among 100 measurements (5.00%)
3 (3.00%) high mild
2 (2.00%) high severe
479.92 µs instead of 938.93 µs. This drops processing time from 833 hours to 415 hours. The moral of the story (somewhat in contrast to the typical software engineering advice): benchmark early, often, and be brutal about narrowly tailored solutions.
Architecture
My basic parallel processing flow looks like:
main()
- Index of Files
- Download File 1
- Process File 1
- Download File 2
- Process File 2
- Download File N
- Process File N
process_file()
- For record in records
- if matches_filter()
- Format json as output
There are two main pipelines in this system with different IO requirements, once we already have the index of warc files that are listed on remote. We first need to download the files from their original source in S3 (network bottleneck). We then need to process these records linearly (CPU bottleneck).
For each record in the file, we need to apply the filtering logic. If it matches our criteria, we can extract whatever metadata is most important to us. We then can concatenate the results to one output file. The simplest convention is 1:1 with the input file so each core is in charge of processing one input file and outputting exactly one output file at a time.
A word on these output files: do some rough estimates on how much data you're expecting to receive. If you copy everything, you're just going to re-derive the original archive files! I squeezed a good bit of extra space with encoding my data into brotli for output. It shrunk output files from ~250MB of mostly html data to ~15MB per output file. For a 1.5GB original file, that's a pretty good data extraction ratio.
Choosing a device
To get through this data in a day, we need a machine with fast pipes to the bucket and as many CPUs as we can get our hands on. So while you don't need to host this in us-east-1, it dramatically speeds up the download times.
per_file = 4min (60s*4)
total_files = 80_000
total_cpu_time = total_files*per_file
per_core_time = total_cpu_time / 192
per_core_hours = per_core_time / 60 / 60
= 27.7h
Final thoughts
My actual execution came in a bit longer than this: at around 30h for the full common crawl, likely due to some of the backoffs and retries when downloading files. This netted out to around 32,000 items processed per second on average (166 per second per core). This 166 covers the decompression, html lexing, language detection, and data extraction on relevant pages.
Rust ended up being the perfect language for this implementation. It lets you very clearly see which operations are copying data or just manipulating a view on-top of it. For implementing some of the low level filtering logic, this clarity is much appreciated. There's also a community focus on optimized packages to really get bare metal performance out of your device. If you have a pretty common task, at this point there's a good chance there's already a rust library that's out there that has been battle tested and performance benchmarked.
There was also something way more enjoyable about this workflow than my previous projects to do the same thing with Spark. Here I spent the majority of my time thinking about algorithmic improvements and memory utilization of the per-record processor, versus trying to sift through remote debugging logs and figure out why the Java worker crashed during a python subprocess spawn. Bugs were reproducible and scaling happened linearly once I switched from local to remote. That might just be personal preference - but I way prefer spending my days battling algorithms than battling remote logs.
The next time I'm working on another big data processing job (at least in the sub-petabyte range), my default is going to remain trying to eek out the most power from one powerful machine. Second to that - deterministically shard over a few of those devices. Turns out one device can take you pretty far.
-
Discovering Common Crawl for the first time in 2014 was one of those core memories as a software engineer. It felt like all the collective intelligence - all the trillions of bits of human experience - were just sitting there, in a public bucket, waiting to be explored.
Of course, I had long known that this information was out there. You can see it every day when you're using a search engine. But indexing the Internet into a static copy is one of these weird engineering problems where it feels so close at hand yet so far away. Scaling up from clicking through links to actually having a redundant archive of billions of pages - with the requisite diversity, deduplication, and robust link frontier - is no small feat. ↢ -
I recognize you could also do a merge with the WAT metadata files that already contain a language tag for each page, but doing this join is non-trivial because of the search space of each index. I also ran into some odd network errors while trying to bulk download the index files. So re-processing the language from the html seemed like the best bet. ↢